Any DASH masternode operators out there?
DASH is currently voting on funding sequencing the cannabis genome to finally close the many remaining gaps using PacBio, Oxford Nanopore, Illumina and Dovetail technology.
If centralized governance won’t fund cannabis research, decentralized governance models may be the more responsive model.
Please weigh in if you feel this is a good use of resources. The genome will be put public with Creative Commons with attribution to DASH.
https://www.dashcentral.org/p/Cracking_the_CannabisGenome_w…
@Dash @CryptoMediaHub @DashForceNews
@Cointelegraph @BitcoinandCryptocurrencyNews
@AllThingsCrypto @SterlinLuxan @BenSwann @RogerVer @ChristopherJ.Carr @ShangoLos @TheCryptoShow @FreeTalkLive @FEE
Cracking the Cannabis Genome on DASH Drive
Tl:Dr
Benefits to DASH
• This is a mechanism to leverage several already funded DASH proposals in the cannabis field that will bring DASH as the main sponsor to CannMed 2018. (https://www.dashcentral.org/p/MedicinalGenomics)
• This will make DASH a permanent name in the Cannabis history and will better leverage the Alt36 presence at many Cannabis conferences, having DASH solve one of the largest scientific challenges in the cannabis field.
• It will make DASH a household name in the Cannabis field and mark an unprecedented event; Cryptocurrencies driving scientific peer review and consensus in a field that can’t get government funding.
• This will also showcase DASH as more than the other Potcoins or hempcoins flooding the cannabis markets.
• It will demonstrate decentralized governance working more effectively than centralized forms of governance.
• It will demonstrate Peer Review and Scientific publishing utilizing the DASH notarizations now available with Stampd.io and crypto-bounty peer review required to move scientific publishing to DASH Drive.
• And it will sequence and publish the most comprehensive Cannabis Genome ever created and for very little investment. We believe this is a unique and productive form of advertisement given recent google, facebook, and youtube bans.
Abstract
DASH has the potential to transform scientific publishing with transparent timestamps, monetization, and censorship resistant publishing. There is a unique opportunity to showcase this potential by taking a highly visible scientific project and leveraging DASH to fund, review, and host the scientific publication of a controversial genome. The ideal genome is the most popular genome that continues to fail to get public funding despite overwhelming public support. We propose to have the DASH network partially fund, peer review and publish the most comprehensive (100X improvement) Cannabis Genome Map ever created and demonstrate why this is an ideal time to make such a move. The economic impact of deciphering the human genome led to a multi-billion dollar human genomics market. We believe the cannabis genome is the next most impactful genome to be completed and decentralized governance may be required to get it done. A better cannabis reference will assist in more rapid molecular breeding programs, better understanding of hermaphroditism, autoflowering genetics and genotype to chemotype predictive markers. It will accelerate more effective breeding and growth of the industry. Demonstrating crypto-incentivized peer review on the DASH network will showcase DASH as leader in the pursuit of truth and censorship resistant publishing. This will be historic.
40 DASH per month for 6 Months.
For best formatting and references use the full Proposal at this link:
https://mega.nz/#!8NZmCAzY!H2h_HfAN0JD6PoOIcoSnbyABwc6WRN8YwG5FehIXLAo
Video tl:dr
Background
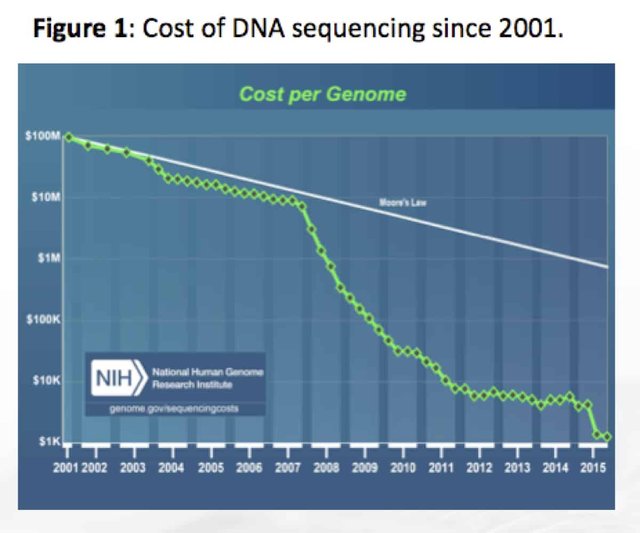
In the past 10 years DNA sequencing technology has decreased in cost nearly 100,000 fold putting Moore’s law to shame. Despite the remarkable achievements in next generation sequencing, the Cannabis genome has remained mostly unreachable to mankind.
In 2011, Medicinal Genomics published the first draft of the Chemdawg Cannabis genome on the Amazon cloud1. Shortly after this publication van Bakel et al published another draft genome for Purple Kush and compared it to hemp and Chemdawg2. These genome references both ended up in over 100,000 pieces (contigs or contiguous sequences) as opposed to the 20 chromosomes it should assemble into. This nonetheless enabled cannabis fingerprinting, marker assisted selection, cannabis microbiome studies, and the hunt for novel terpenoid synthase genes2-7. Despite the progress enabled with a draft reference, a fully contiguous genome assembly is required to perform epigenetic studies regarding the mechanisms of hermaphroditism, autoflowering genetics, pest resistance and a better understanding of the expression of rare cannabinoids.
The cannabis genome assembly challenge is largely due to the fact that the diploid genome is 65% AT and over 1% polymorphic. For reference, the human genome is 56% AT and has a polymorphism rate 10 fold lower (0.1%). The polymorphism rate is the rate at which the maternal and paternal genomes differ. Each cell in the plant has 2 genomes mixed (one from the mother and one from the father) and the process of isolating DNA can’t tease these apart. As a result one is left with 2 near identical genomes mixed together for sequencing. Since one cannot read telomere to telomere (from the end of one chromosome to the other) with current DNA sequencing technologies, DNA molecules are randomly broken up into smaller but overlapping pieces and read redundantly. This is a process known as Shotgun Sequencing (https://en.wikipedia.org/wiki/Shotgun_sequencing). With enough overlapping molecules, the sequence can be read and reassembled computationally. This is where read length becomes critically important as the longer the read, the more signature you have and the more overlap can be found in repeat rich, entropy poor genomes like Cannabis.
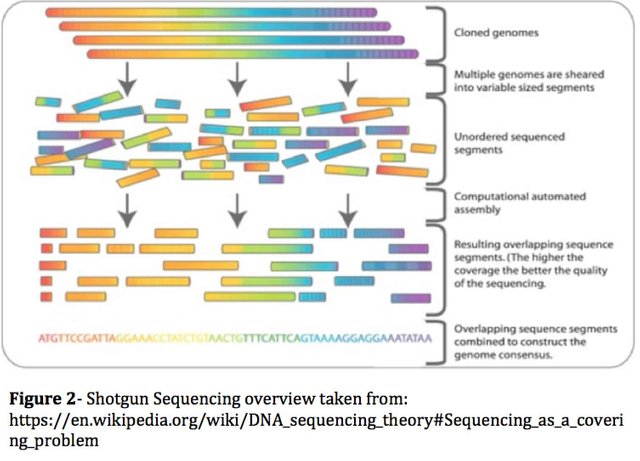
To further illustrate the computational complexity, imagine a billion piece jigsaw puzzle where 65% of the pieces are Red and Green and where it has been mixed with another puzzle of near identical nature with the expectation of 1 puzzle piece out of 100 being altered. DNA assembly is the job of sorting those two puzzles out into their own respective correct assemblies. For 800 Megabase (Mb) genomes it remains a computationally difficult task in 2018.
This extreme base imbalance combined with a highly polymorphic genome has evaded improvement since 2011. While a few attempts at using low coverage Pacific Bioscience sequence have been submitted to NCBI, those attempts appear to be missing 25% of the genome (only 550Mb where 2011 assemblies point to 750Mb total genome size). These later assemblies do showcase longer contiguity as expected with the longer read length platforms employed. There have also been a few “science by press release” suggestions of other companies working on this but no data has been publicly released to verify these claims.
We believe the technical solution to this problem is to use multiple sequencing platforms to error correct and expand the assembly (PacBio, Illumina, MinION), but also higher depth Pacific Bioscience sequencing (50X) and multiple Dovetail libraries to span long chromosomal segments together. Medicinal Genomics owns both MinION and Illumina sequencing platforms and currently outsources the Pacific Bioscience sequence. DNA sequence data from all 3 platforms has already been contributed to the project (see links below). The Dovetail libraries have never been deployed on Cannabis and we believe these novel libraries combined with the more modern long read platforms will bring the highest resolution cannabis map ever created8, 9.
The failure to assemble a better cannabis reference has occurred despite the most exciting improvement in read length chemistry in DNA sequencing since its invention by Fred Sanger in 1977. Since the invention of DNA sequencing, read lengths have been limited to 1000 base (bp) reads for 40 years. In 2013, Pacific Biosciences and Oxford Nanopore began making revolutionary improvements with single molecule DNA sequencing by pushing the read lengths out to 40,000bp and 400,000bp respectively. While these read lengths were impressive, they were largely achieved by using noisy single molecule sequencing techniques (10-20% error rates). In 2018 these long read platforms are beginning to improve their raw read accuracy to a point where complex plant genomes are now within reach. The accurate long read sequencers have fundamentally changed the face of DNA sequencing. It is the most disruptive achievement in genomics history next to the sequencing of the human genome itself.
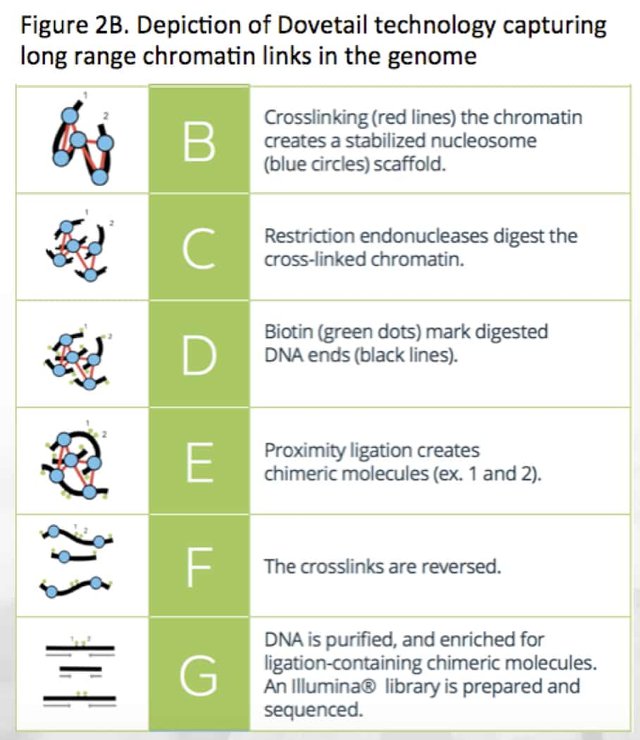
These longer read platforms do have some limitations. The most significant one relates to the length of DNA one can purify from a cell. Manual pipetting often hydrodynamically shears DNA into 30,000bp fragments. The most careful techniques have achieved 1Mb reads but to get 10Mb to 100Mb information from DNA, chromatin cross-linking libraries like Dovetail Chicago libraries are required (Figure 2B). The author has published experience generating nucleosome positioning maps with Nobel Laureate Dr. Andrew Fire10.
Applications of these technical improvements to cannabis genomics have been slow to mature primarily because most labs that can afford such sequencers have federal overhangs that prevent them from working on a federally illegal plant. Likewise, expensive projects that are simply put public don't always attract private businesses to fund them to completion. If start-ups don't see immediate commercialization of such a public resource, the investors usually tire of the academic trophy pursuit and ask for the entrepreneurs to focus on more immediate products. Private cannabis companies can also be difficult to fund in the cannabis space due to federal banking regulations making the cannabis genome project one that has stalled for 7 years during DNA sequencings most impressive improvements.
To date, Medicinal Genomics has funded over $300,000 worth of public genomic sequencing in Cannabis and we have been productively organizing others to contribute to the public domain. We have a very strong open source ethos and have even funded other commercial competitors in the space to make their data public. The Lynch et al paper is an example of multiple small and potentially competitive startups in the cannabis space contributing data, time and money to making many cannabis genotypes public. I personally funded the open access fee for this manuscript and many genomic sequences used in the study.
Today, the field of cannabis genomics is mostly utilizing the more affordable short read sequencers (Illumina) to perform “resequencing” of genomes to fingerprint the plants for defensive IP purposes. Kannapedia.net was built to leverage this. All of the data on Kannapedia.net would be vastly improved if mapped against a much more contiguous and complete cannabis reference sequence. We experimented with all public cannabis references available in 2017 and the references from 2011 still map the most reads and are what is currently being used in Kannapedia.net.
Kannapedia.net notarization was recently migrated to the DASH blockchain thanks to another DASH proposal and we hope to complete this migration by having the most complete reference genome be DASH funded and hosted. This DASH notarization migration occurred within 30 days of DASH funding and has been noticed in the cannabis community11, 12. DASH is the lead sponsor for CannMed 2018 and this will be a prime conference to reveal progress on the assembly.
To address this scientific challenge, we propose the utilization of the DASH network to drive the assembly of an improved Cannabis Genome reference in a manner that has multiple other parties with “Skin In The Game”. This Project will demonstrate DASH cryptocurrency can fund critical and popular medically relevant science where the political system will not. This will showcase cryptocurrency as being responsive to popular scientific demands and in a manner that demonstrates more distributed storage on DASH Drive than what is traditionally seen at NCBI and other tax run genomic databases. Current databases at NCBI are prone to government shutdown during budget ceiling debates and there is reasonable apprehension in the cannabis field using this as an exclusive store of Cannabis data. Databases that have a pricing signal are also more disaster resistant. While we don't object to using these public resources, we believe there is good reason to mirror these data in other places given the current administrations change in opinion on Cannabis. Given the genome sequence materializes faster than DASH Drive, we have placed contingency hosting funds on MEGA.nz in the budget.
The project will also demonstrate that the scientific peer review process can be dis-intermediated with cryptocurrency. By utilizing crypto incentivized peer review (RAIDreviews.org), we can dis-intermediate pay wall journals and the tired copyright system. These crypto-bounty based audits on the project also enhance transparency and accountability with the DASH network. We propose two such audits for this project where $1000 crypto bounty will be offered to run peer review diagnostics on the state of the assembly.
The project can be incrementally monitored for success or failure and will provide high visibility of the DASH DAO to the scientific community. We believe this is increasingly important as Facebook, Google, YouTube and other entities ban crypto advertisements.
The best advertisement is the work you stand up for. Leading by example speaks louder than ‘ad’ dollars on a surveillance platform. Instead of Crypto being blamed for the drug trade, turn the tables and demonstrate that crypto is actually on the right side of history funding the very science needed to advance the public discussion. Cannabis legalization has been more popular than any of the last 3 US presidents. When people see a currency that reacts to their opinion faster than the political system can react with fiat monetary policies, they may begin to question the legitimacy of their monetary system and look to DASH as a solution. The cannabis industry is a receptive audience to more just alternatives than the unconstitutional banking coercion they struggle with everyday.
Benefits to the Community
Recent genomic surveys on cannabis have demonstrated gene duplications in the genes of most medical interest. Single molecule sequencing of THCA Synthase implies 4-6 copies THCA synthase exist in many Type I cannabis genomes 13-15. Likewise, many of the genes in the terpenoid and cannabinoid synthesis pathway appear replicated and hyper-polymorphic. This is unsurprising given the intense selective pressure placed on these genes. Breeders have been selecting for increased terpenoid and cannabinoid expression since prohibition began prosecuting on total plant weight and this selection has resulted in plants with replicated gene content in these pathways. A perfected cannabis reference genome would resolve these duplication events into their precise chromosomal locations and enable the classification of variants that drive higher chemical expression.
The main economic benefit of a better cannabis reference is related to the price of DNA sequencing. Long Read technologies required to build the first map of cannabis can be 100X more expensive than shorter read resequencing platforms. Once the human genome was in less than 1000 pieces, cheaper short read sequencers could be used to scan for most of the clinical utility. This is reported to have created 310,000 jobs and over $796B of economic gain16. While this report suffers from some Kaynesian logic a more conservative estimate is the anticipated annual $26B DNA Sequencing market in 2025 growing with a 20% CAGR. The current DNA sequencing market is mostly human sequencing with Oncology, Clinical investigation, reproductive health and organ transplant HLA typing taking over 75% the total DNA sequencing market17.
This makes the much more affordable target capture and resequencing efforts available on the human genome, far more challenging and less effective in the cannabis genome today. Building a better public reference would bring all researchers in the cannabis space the capacity to more effectively utilize much cheaper short read sequencers and help to decentralize cannabis genetics. Likewise, resolving these medically relevant copy number alterations would help link genotypes to predicted chemotypes and enable many more researchers to better annotate these important medicinal pathways.
Strategy and Methods
High Molecule Weight (HMW) DNA purification is critical to enabling long read sequencers. Your DNA read length is limited to the size of the molecules you present the long read sequencers. We have perfected DNA purification methods in collaboration with New England Biolabs and they have already run ~10Gb of Pacific Biosciences sequence on the Blueberry Cheesecake genome (link below). This was a voluntary contribution to the project and demonstrates collaborators with substantial “skin in the game”. Another 35Gb of Pacific Bioscience sequence is needed and the DNA is ready to go once funding is in place. These methods usually produce 10-30kb reads that need to be assembled into a draft assembly.
The Blueberry Cheesecake genome was selected as it is a hermaphrodite prone Type II cannabis plant that is Terpinolene and Beta caryophyllene dominant. To date, cannabis sequencing efforts have primarily focused on THCA dominant Type I plants (Chemdawg, LA Confidential, Purple Kush) and CBDA dominant Type III plants (Cannatonic, USO-31, Finola), while Type II plants have been the subject of controversial cannabis patents. Terpinolene has published benefits in anxiety and cancer18, 19. Beta caryophyllene is often characterized as a non-cannabis exclusive cannabinoid with CB2 receptor affinity20, 21. For more information regarding Cannabis plant type nomenclature see “The inheritance of chemical phenotype in Cannabis sativa L”.22.
The other critical factor in sequencing a plant genome, is access to milligrams quantities of purified DNA. We have this DNA already purified. The moment you run out of DNA, your sequencing project is over. As a result it is critical to have access to excessive plant DNA to make multiple long read libraries.
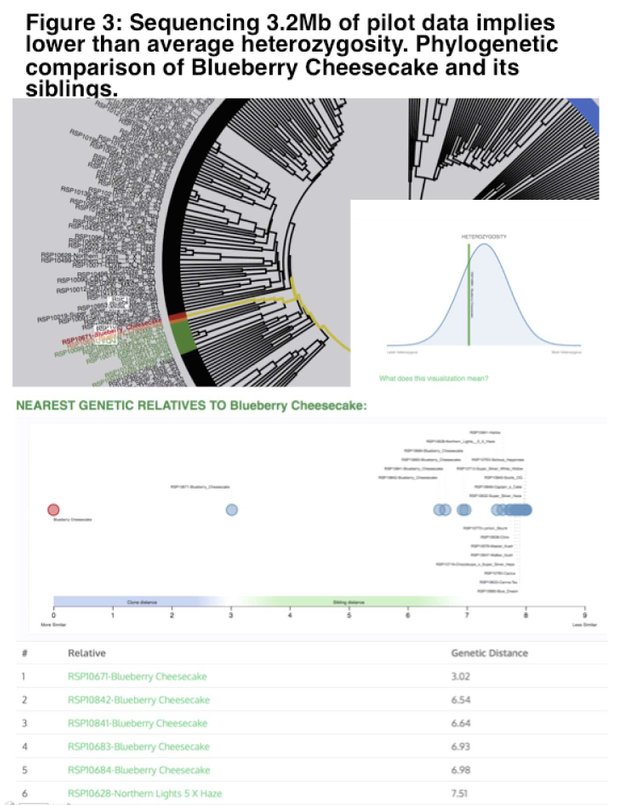
Additionally, small amounts of DNA are available from multiple siblings of this plant. These have been preliminarily sequenced with Medicinal Genomics StrainSEEK system. This test sequences 820,000bp – 3.2Mb in these plants to establish a fingerprint for phylogenetic comparison. By doing so we can see that these lines are more homozygous than the average sample we tend to sequence for customers. Assemblies are most successful with less heterozygous genomes.
Draft Assembly
DNA assembly is a RAM intensive process. For complex plant genomes, over 512Gb RAM is usually required. To achieve this we utilize AWS x1.32xlarge memory optimized compute cluster at $13.38/hour. Each assembly attempt is expected to take 336 hours on this platform or $4500 per attempt. The Canu assembler was chosen based on its publish success but also simplicity in install and assembly monitoring features. It is important to have Canu real time feedback on compute jobs that run for days or weeks at $13/hour. Canu also enable “restart from” command line control of the process, greatly limiting Compute costs. Dr. Harkins is listed as an assembly consultant at $500/hour. We estimate 3 hours of work for every assembly setup and attempt. Five incremental assemblies are budgeted for. Dr. Harkins can accept DASH directly. Dovetail will also be attempting an Assembly improvement with their HiRise software.
Dr. Harkins resume:
The Canu Assembler
Amazon Pricing
Assembly polishing
Single molecule sequencers often produce single nucleotide insertion or deletion errors (indels) that can be polished with highly accurate short read sequencer like the Illumina HiSeq. Medicinal Genomics will provide HiSeq data to help error correct the Pacific Biosciences long read data. In addition to indel polishing, repeat structures longer than the 30Kb readlength will leave branches or forks in the assembly graph. To resolve these repeat structures, Dovetail libraries will be made. These libraries leverage chromatin cross-linking and can scaffold genomes into Megabase sizes often providing full chromosome assemblies. These libraries are expensive to make and will be approached incrementally to constrain costs.
Dovetail Hi-C Technology
Project success and monitoring
Monitoring the success of DASH proposals can be challenging in highly specialized fields. To address this we propose a mid-project crypto bounty incentivized peer review be performed with the project. These RAID reviews (http://raidreviews.org) can be completed in under a week and provide valuable independent insight to the DASH community on the status of a given project. In the case of the pilot review for the Kratom genome project, 0.1BTC was used to incentivize a 4 day peer review of genome assembly status. In addition to a mid project peer review, we plan to follow the spirit of the Bermuda Principles in adopting rapid public data release of all raw DNA sequence generated thus enabling even our competitors to pick up the data and compete with us. Pre-Print publications may be pursued in addition to full publication in a respected journal. Only open access journals will be considered and the Crypto-Bounty peer review process can be compared to the more traditional methods. Traditional methods may take 4-5 months compared to the weeks seen in the crypto-bounties projects. I will also be speaking at 2 or more conferences before CannMed and would plan to speak on this topic. One talk is at The Broad Institute in Cambridge MA. Another presentation is at the Society for In vitro-Biology.
The Broad Institute Meeting
SIVB Meeting
Risks
There is risk that this genome assembly is harder than we are anticipating but we have significantly derisked this after 7 years of experience assembling various cannabis genomes and other model genomes. We have more experience in Cannabis genomics than any team on earth and believe this is the right strategy and the technology is finally here to do it right.
There is risk others will publish an assembly first. There is evidence of 3-4 other small private laboratories attempting to improve the genome reference sequence (Phylos, Steephill labs, Anandia labs, SunRise genetics). All are placing their DNA sequencing efforts in government databases that go offline every time there is a government budget crisis. None of these labs have government funding. Very few own their own DNA sequencers. As a result, their efforts are likely limited by the capacity to move plant tissue to various service providers required to make this project mature. These labs are mostly focused on sequencing either Type I or Type III plants. No one to date is sequencing a Type II plant (expresses both THCA and CBDA).
Even if these labs succeed, the field will require more than one Cannabis reference genome and the cross comparisons of each dataset will be highly valuable. It is likely Type I-V cultivars will each need their own reference sequence. We suspect the industry will also sequence hemp cultivars for seed and fibre production. Ruderalis and landrace sativa cultivars will likely be sequenced just to understand the historical genetics that may have been lost due to prohibition. Likewise, male, female and hermaphroditic genomes will eventually be sequenced. In summary, the field is so young and desperate for data that there is limited risk of being ‘scooped’. A collaborative comparison study is more likely to emerge than a zero sum game.
The process of having this funded, reviewed and stored by a cryptocurrency blockchain is unique and will make a resounding impression in public opinion regarding cryptocurrencies delivering the truth in scientific fields where the political process has failed us.
Pilot Goals
Improve the Cannabis Genome Reference contiguity 100 Fold (N50s of 2Kb to 200Kb)
Pacific Bioscience sequence coverage of 50X should deliver 50-100Kb N50s.
Dovetail Chicago Libraries should increase contiguity beyond 200Kb.
Presentation of project status at CannMed 2018, Oct 22-24 (DASH is a Sponsor).
Costs
For major item we have provided links to the quotes from various providers.
35Gb of Pacific Biosciences Sequel data ($31,300)
PacBio Quote
HWM DNA Purification Amplicon Express ($3,550)
Amplicon Express Quote
Dovetail Chicago and HiC Library generation & High Rise Assembly($11,450)
Dovetail Quote
Dovetail Illumina Library Sequencing at Medicinal Genomics ($25,000)
DNA Assembly using Canu at AWS ($30,000 or $6,000 x 5 attempts)
Amazon Pricing
Optional Oxford Nanopore scaffolding with MinION 1D^2 reads ($10,000)
RAID review (2x $1000)
Final Peer Review and Publication ($3500)
Contingency Data Hosting Fees in the event DASH Drive isn’t ready ($1500)
5 DASH Proposal fee ($2550)
Travel and consulting ($550)
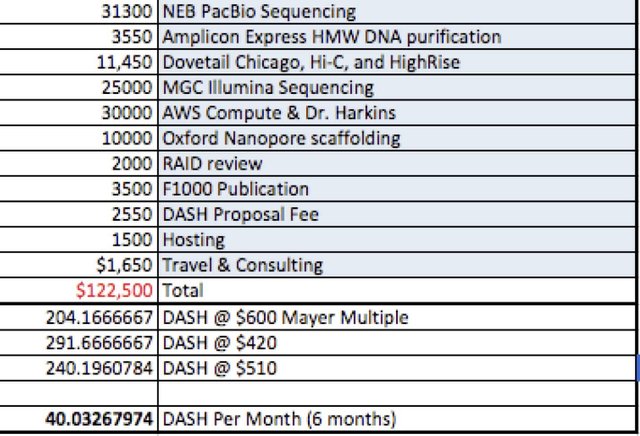
$122,500 At $510/DASH = 240 DASH or 40 for 6 months.
We propose a DASH price that is halfway between the DASH Mayer Multiple of $600USD/DASH and current day price of $420 to derisk currency volatility.
DASH Mayer Multiple
Future Directions
We envision a future proposal to encompass the build of a more scalable genome browser than Kannapedia.net. Upgrades to this would include more decentralized store of the data, with higher searchability and better genome annotation viewers. Until more information regarding the finished genome size and DASH Drives architecture are in hand this is difficult to price into todays work. Preliminary estimates are in $60K range but we are still scoping and quoting this aspect of the work.
Donations already made by collaborators with Skin in The Game
This proposal marks the first time this Cannabis genomic data has ever been placed public. We have also attached a Case Study where this strategy was used to improve a more complex genome than Cannabis (Plantain genome). This case study implies a very high likelihood of success for the strategy selected.
10Gb of Pacific Biosciences sequence from NEB
https://mega.nz/#!dZpHECbQ!MQGCf0AcQ3g-LyzlnmWsjdbLWgLeg_X0x1Q8CAvB2B0
https://mega.nz/#!4Mx0mDLb!_FQ2CA13nmzu8w4ieK7MEgbtcMqwMp8MYkWCCAcF4Y0
6 StrainSEEKs of Blueberry Cheesecake siblings
http://www.kannapedia.net/strains/rsp10573/
http://www.kannapedia.net/strains/rsp10684/
http://www.kannapedia.net/strains/rsp10682/
http://www.kannapedia.net/strains/rsp10683/
http://www.kannapedia.net/strains/rsp10670/
http://www.kannapedia.net/strains/rsp10671/
[b][i]1 flow cell of Oxford Nanopore MinION data (1Gb)[/i][/b]
[b]Case Study on the Plantain Genome Project[/b]
Recipient track record
Medicinal Genomics was a fortunate recipient for a small DASH grant that has successfully been used to enable Kannapedia.net to migrate its Genomics notarization service over to the DASH blockchain. This was achieved with Stampd.io in 30 days from funding. The other DASH funds are being successfully utilized in CannMed 2018 sponsorship. These funds have enabled successful organization of an outstanding speaker list and further recruitment of interested sponsors. We currently have a track record of being very responsive to DASH inquiries and delivering on goal.
DASH-CannMed 2018 Headline Speakers
The lead organizer of this proposal was the Team Leader for R&D at the Whitehead Institute/MIT center for genome research. He is an author on the Human Genome Project23 and well published in the DNA sequencing field having invented and developed two of the Next Generation Sequencing platforms (SOLiD and Ion Torrent)24-26. Tim Harkins, PhD. is an assembly adviser to the project and is equally well published in the genomics field with focus on long read DNA sequencing technology development and utilization.
[b]My biography can be found here:[/b]
- Stafford. Sequencing the Cannabis Genome: Impact, History, and Future. American Botanical Council. 2011;8(10).
- van Bakel H, Stout JM, Cote AG, Tallon CM, Sharpe AG, Hughes TR, et al. The draft genome and transcriptome of Cannabis sativa. Genome biology. 2011 Oct 20;12(10):R102. PubMed PMID: 22014239. Pubmed Central PMCID: 3359589.
- McKernan K, Spangler J, Helbert Y, Lynch RC, Devitt-Lee A, Zhang L, et al. Metagenomic analysis of medicinal Cannabis samples; pathogenic bacteria, toxigenic fungi, and beneficial microbes grow in culture-based yeast and mold tests. F1000Research. 2016;5:2471. PubMed PMID: 27853518. Pubmed Central PMCID: 5089129.
- McKernan K, Spangler J, Zhang L, Tadigotla V, Helbert Y, Foss T, et al. Cannabis microbiome sequencing reveals several mycotoxic fungi native to dispensary grade Cannabis flowers. F1000Research. 2015;4:1422. PubMed PMID: 27303623. Pubmed Central PMCID: 4897766.
- McKernan KJ. The chloroplast genome hidden in plain sight, open access publishing and anti-fragile distributed data sources. Mitochondrial DNA Part A, DNA mapping, sequencing, and analysis. 2016 Nov;27(6):4518-9. PubMed PMID: 26486305.
- Booth JK, Page JE, Bohlmann J. Terpene synthases from Cannabis sativa. PloS one. 2017;12(3):e0173911. PubMed PMID: 28355238. Pubmed Central PMCID: 5371325.
- Lynch. Genomic and chemical diversity of cannabis. Critical Reviews in Plant Sciences. 2016;35(5-6).
- Research signals arrival of a complete human genome. PHYSORG.https://phys-org.cdn.ampproject.org/c/s/phys.org/news/2018-03-human-genome.amp.
- Jain M, Olsen HE, Turner DJ, Stoddart D, Bulazel KV, Paten B, et al. Linear assembly of a human centromere on the Y chromosome. Nature biotechnology. 2018 Mar 19. PubMed PMID: 29553574.
- Valouev A, Ichikawa J, Tonthat T, Stuart J, Ranade S, Peckham H, et al. A high-resolution, nucleosome position map of C. elegans reveals a lack of universal sequence-dictated positioning. Genome research. 2008 Jul;18(7):1051-63. PubMed PMID: 18477713. Pubmed Central PMCID: 2493394.
- Amirault. BLOCKCHAIN TECHNOLOGY IS A MODERN REQUIREMENT FOR THE CANNABIS INDUSTRY. Growers Network. 2018;http://growersnetwork.org/market-projections/blockchain-technology-modern-requirement-cannabis-industry/.
- Scott. Bitcoin 'Too Slow' for cannabis industry until block size changes. Bitcoinistcom. 2017;http://bitcoinist.com/bitcoin-slow-cannabis-block-size/.
- Weiblen GD, Wenger JP, Craft KJ, ElSohly MA, Mehmedic Z, Treiber EL, et al. Gene duplication and divergence affecting drug content in Cannabis sativa. The New phytologist. 2015 Dec;208(4):1241-50. PubMed PMID: 26189495.
- Onofri C, de Meijer EPM, Mandolino G. Sequence heterogeneity of cannabidiolic- and tetrahydrocannabinolic acid-synthase in Cannabis sativa L. and its relationship with chemical phenotype. Phytochemistry. 2015 Aug;116:57-68. PubMed PMID: 25865737.
- Kevin J McKernan YH, Vasisht Tadigotla, Stephen McLaughlin, Jessica Spangler, Lei Zhang, Douglas Smith. Single molecule sequencing of THCA synthase reveals copy number variation in modern drug-type Cannabis sativa L. BioRXIV. 2015.
- Tripp. Economic Impact of the Human Genome Project. https://wwwbattelleorg/docs/default-source/misc/battelle-2011-misc-economic-impact-human-genome-projectpdf?sfvrsn=6. 2011.
- Research GV. Next Generation Sequencing (NGS) Market Size & Forecast By Application https://wwwgrandviewresearchcom/industry-analysis/next-generation-sequencing-market. Jan, 2017.
- Aydin E, Turkez H, Tasdemir S. Anticancer and antioxidant properties of terpinolene in rat brain cells. Arhiv za higijenu rada i toksikologiju. 2013 Sep;64(3):415-24. PubMed PMID: 24084350.
- Okumura N, Yoshida H, Nishimura Y, Kitagishi Y, Matsuda S. Terpinolene, a component of herbal sage, downregulates AKT1 expression in K562 cells. Oncology letters. 2012 Feb;3(2):321-4. PubMed PMID: 22740904. Pubmed Central PMCID: 3362481.
- Legault J, Pichette A. Potentiating effect of beta-caryophyllene on anticancer activity of alpha-humulene, isocaryophyllene and paclitaxel. The Journal of pharmacy and pharmacology. 2007 Dec;59(12):1643-7. PubMed PMID: 18053325.
- Gertsch J, Leonti M, Raduner S, Racz I, Chen JZ, Xie XQ, et al. Beta-caryophyllene is a dietary cannabinoid. Proceedings of the National Academy of Sciences of the United States of America. 2008 Jul 1;105(26):9099-104. PubMed PMID: 18574142. Pubmed Central PMCID: 2449371.
- de Meijer EP, Bagatta M, Carboni A, Crucitti P, Moliterni VM, Ranalli P, et al. The inheritance of chemical phenotype in Cannabis sativa L. Genetics. 2003 Jan;163(1):335-46. PubMed PMID: 12586720. Pubmed Central PMCID: 1462421.
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, et al. Initial sequencing and analysis of the human genome. Nature. 2001 Feb 15;409(6822):860-921. PubMed PMID: 11237011.
- McKernan KJ, Spangler J, Helbert Y, Zhang L, Tadigotla V. DREAMing of a patent-free human genome for clinical sequencing. Nature biotechnology. 2013 Oct;31(10):884-7. PubMed PMID: 24104751.
- McKernan KJ, Peckham HE, Costa GL, McLaughlin SF, Fu Y, Tsung EF, et al. Sequence and structural variation in a human genome uncovered by short-read, massively parallel ligation sequencing using two-base encoding. Genome research. 2009 Sep;19(9):1527-41. PubMed PMID: 19546169. Pubmed Central PMCID: 2752135.
- Rothberg JM, Hinz W, Rearick TM, Schultz J, Mileski W, Davey M, et al. An integrated semiconductor device enabling non-optical genome sequencing. Nature. 2011 Jul 20;475(7356):348-52. PubMed PMID: 21776081.
This topic is super hard for me to understand
but thanks for the info :)
Downvoting a post can decrease pending rewards and make it less visible. Common reasons:
Submit
Following voted and shared! This is why I am here. I can support and help grow these kinds of projects!
Downvoting a post can decrease pending rewards and make it less visible. Common reasons:
Submit
You're touching a lot of important points here, botanically, genetically, economically and methodologically, but disintermediation of peer-review process with cryptocurrency is probably one of the most audacious ones :)
I'd love to see full genome deciphered to "perform epigenetic studies regarding the mechanisms of hermaphroditism, autoflowering genetics, pest resistance and a better understanding of the expression of rare cannabinoids."
But I'd say we also need it to start modern breeding programmes for stability and for outdoor growing in different climates. We should select the most homozygous lines in the process and these should become our P1 stock.
Well exciting times ahead!
Downvoting a post can decrease pending rewards and make it less visible. Common reasons:
Submit
Check out KratomDNA.org and RAIDreviews.org.
Downvoting a post can decrease pending rewards and make it less visible. Common reasons:
Submit
Can't read the entire thing but fully support your work. Resteeming this now! Voting and as soon as I set up auto-vote you will be on it. And also have the support of @ganjafarmer ! This is huge! We can preserve the original strains forever and more research!
You always have our votes! Please keep this essential work up
Downvoting a post can decrease pending rewards and make it less visible. Common reasons:
Submit
We are sequencing a TypeII plant described in the recent batch of 4 cannabis patents from Biotech LLC. The patents do not cover the DNA but I am explicitly doing this to drive more awareness at the USPTO. Once we have genomes of TypeII plants, Anyone can sequence a strain from before 2013 and if it matches this, it will invalidate those patents!
Downvoting a post can decrease pending rewards and make it less visible. Common reasons:
Submit
Keep advancing the science of the healing plant! Love it!
Downvoting a post can decrease pending rewards and make it less visible. Common reasons:
Submit
Its back up for Vote- Looking much better this time with a 50% reduction in the budget.
Ernest Hancock also suggested I get a DASH address so others can chip in and be on the credits page.
https://www.medicinalgenomics.com/crypto-funded-public-genomics/
https://www.freedomsphoenix.com/Media/Media-Files/001-0522110515-2018-05-22-Hour-2-Kevin-McKernan---MedicalGenomicsCOM---DASH.mp3
Here is the proposal -
https://www.dashcentral.org/p/MedicinalGenomics_II
Thanks for the support
Downvoting a post can decrease pending rewards and make it less visible. Common reasons:
Submit