Along with many fascinating features of the cannabis plant and human uses thereof, issues around the strain-name game have recently been brought up to the surface, especially in the wake of the impending legalization of cannabis for recreational use.
Given marked advances in DNA sequencing technology and the newly available draft cannabis genome, a strong push to end the strain-name game seems to be associated with a sizably larger industry, prone to piracy and all the other things that happen when a market goes mainstream. Indeed with more demand comes more supply, and given that recreational regulations will likely not meet the stringent criteria of established medical laws, companies and regulators alike are in need of replicable high-throughput methods to differentiate plants of divergent origins.
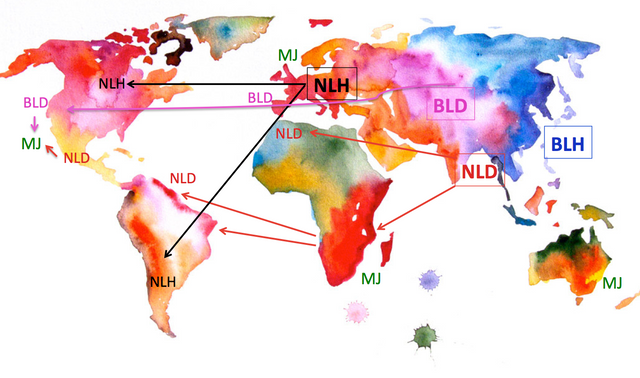
While it is well accepted that the Cannabis genus includes four main gene pools, which I like to refer to as sub-species: Narrow Leaf (European) Hemp (NLH; C. sativa sativa), Broad Leaf (Chinese) Hemp (BLH; C. sativa chinensis), Narrow Leaf Drug-type (NLD; C. sativa indica) and Broad Leaf Drug-type (BLD; C. sativa afghanica), the clustering of accession within each of these sub-species has only recently began to gain interest from the scientific community (Sawler et al 2015, Henry, 2015, Lynch et al 2015). Future large scale sampling, particularly of "landrace" samples may reveal further affinity between geographically, politically and historically connected populations that may warranty further taxonomic revisions.
Current endeavours have yielded some congruent and contrasting patterns, but in a general sense, all authors agree on the fact that cannabis strains found in the current medical and recreational markets in North America (referred to from here on as MJ strains) are extensive hybridized plants with NLD and BLD ancestry with high CBD varieties incorporating some portion of the pool of the European and/or Chinese Hemp. Another key finding pertains to the large amounts of genetic variation found in MJ strains compared to European hemp varieties, leading some to speculate as to the role that prohibition has played in controlling breeding efforts.
One has to acknowledge at this point that the goal of selective breeding is to obtain a true breeding population, which by definition is homozygote at the traits of interest. Highly selected true breeding populations should thus have lower genetic variation. It is likely that with cannabis coming out of the dark ages the tools of genomics will be harnessed in selective breeding programs and we will see the levels of genetic variation within individual MJ strains get selectively lower as we unleash the power of genomic selection in breeding programs for particular traits of interests, particularly related to therapeutic potentials. This idea of course is not novel and was accepted for publication on my 23rd birthday, over ten years ago (Pacifico et al 2006).
Within the scope of 21st century Cannabis in North America, understanding the phylogenetic relationships among different MJ strains will help shed light on the origin or particular traits of interest, their interrelationships, and will yield useful tools to establish prior art in terms of novel hybrid combinations. Indeed, several groups have recently undertaken this endeavour, with the goal to provide a time stamp on the unique genetic makeup of a large number of MJ hybrids, and some open initiatives are currently available at http://www.kannapedia.net/ and http://galaxy.phylosbioscience.com/.
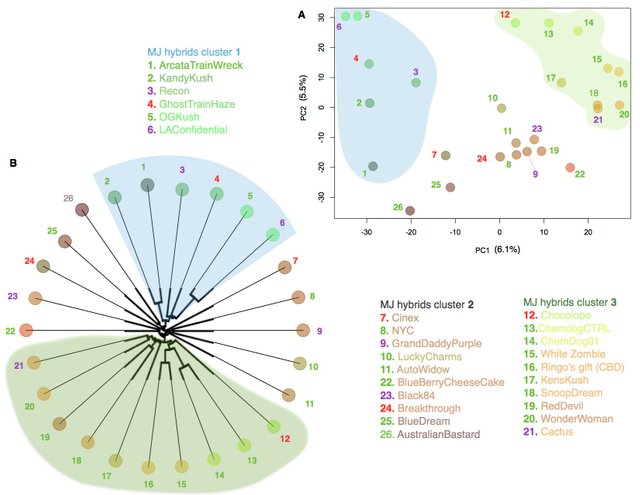
While Medicinal Genomics provides open access to their data for anyone to download and analyse, Phylos has currently not released their data, but intend to do so through their Open Cannabis Project. Thus in the meantime, let us take a look at the clustering of MJ hybrids from a dataset of 26 MJ hybrids typed at over 180’000 single nucleotide polymorphisms (SNPs):
Two separate means of depicting the relationships between strains was used, principal component analyses (PCA) and phylogenetics illustrated in an unrooted tree. From both approaches, one can discern three main clusters in MJ hybrids with no particular affinity for either Indica, sativa or hybrids epithets, although cluster 3 was made up mainly of hybrids.
Of interest to the strain-name game, two accessions were found to be overlapping in the PCA: Snoop’s Dream and Cactus, making them good initial candidates to resolve the game and establish a nomenclature consensus. While these analyses are coarse as the dataset contains a very large number of SNPs that may not all be informative, this brief note is intended to demonstrate the utility of applying genomic tools to establish a precedent in terms of the importance of identifying genetically unique accessions and recognising them as such. This research will greatly benefit from the incorporation of a larger number of accession, which may be forthcoming in the years to come. In the meantime, I hope those with unique genetics will approach one of the genetic providers in the cannabis space to submit their samples in order to complete this emerging picture.
This Story was originally posted on Canlio.com and can be retrieved here
References:
Henry P. Genome-wide analyses reveal clustering in Cannabis cultivars: the ancient trilogy of a panacea. PeerJ PrePrints; 2015 Nov 30.
Lynch RC, Vergara D, Tittes S, White K, Schwartz CJ, Gibbs MJ, Ruthenburg TC, Land DP, Kane NC. Genomic and Chemical Diversity in Cannabis. bioRxiv. 2015 Jan 1:034314.
Pacifico D, Miselli F, Micheler M, Carboni A, Ranalli P, Mandolino G. Genetics and Marker-assisted Selection of the Chemotype in Cannabis sativa L. Molecular Breeding. 2006 Apr 1;17(3):257-68.
Sawler J, Stout JM, Gardner KM, Hudson D, Vidmar J, Butler L, Page JE, Myles S. The genetic structure of marijuana and hemp. PloS one. 2015 Aug 26;10(8):e0133292.
Hi! I am a content-detection robot. I found similar content that readers might be interested in:
https://www.canlio.com/blog/cannabis-genetics#!
Downvoting a post can decrease pending rewards and make it less visible. Common reasons:
Submit