Definition of life
The size of life spans seven orders of magnitude, from invisible bacteria to blue whales. However, there is not a universally accepted definition of life due to its complexity and versatility. In fact, many definitions choose to describe the fundamental properties of living organisms as a prerequisite to life. According to NASA, ‘Life is a self-sustaining chemical system capable of Darwinian evolution’. Their definition breaks down into two branches, specifically metabolism and replication. However, these two features combined result in a highly complex system, making the transition from non-life a low probability event. Therefore, an intermediate step is required which would identify the boundary between living organisms and inorganic life. In reality, this process was a cascade of transition states (4), which were driven by catalysts to overcome successive activation energies. The actual kinetics behind this open system pose the biggest unsolved mystery in the origin of life. In addition, the question of whether or not metabolism preceded replication remains unanswered, although there are experiments that support the latter (5), equating the origin of life to the origin of first replicators.
Due to recent advancements in technology, especially computer science and more importantly artificial intelligence, existing definitions will be too narrow to consider all aspects of life. In order to narrow down the scope of this report, the main focus will be the origin of cellular life on Earth, based on the various existing hypotheses.
Life’s building blocks include proteins, carbohydrates (sugars), lipids and nucleotides, all of which are made of long chain molecules. This field studies the transformation of prebiotic chemistry into vesicle membranes, RNA molecules and peptides through polymerisation reactions. Impressively, the first law of thermodynamics which states that energy is conserved in an isolated system is obeyed in biological systems too, whereby all living organisms contain a genome; the essential core of life. The two key molecules that are conserved across all known life forms are shown in figure 1, which aims to illustrate their composition and 3-D shape. The highly complex structure, coupled with intermolecular forces acting with the surrounding system leads to fundamental properties of living organisms.
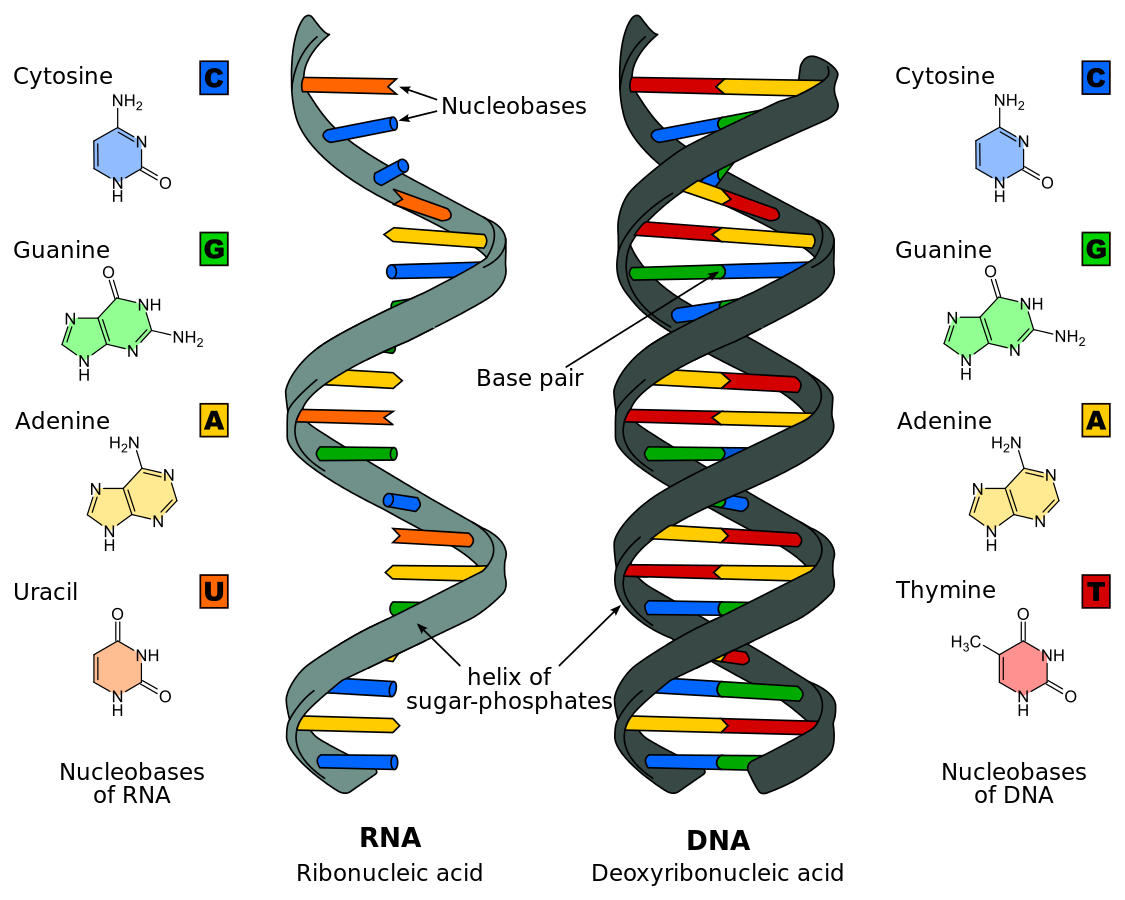
Figure 1: Comparison of a single-stranded RNA and a double-stranded DNA with their corresponding nucleobases. Image courtesy of Roland
Initially, the hypothesis of spontaneous generation lasted for almost 2 millennia and was only disproven after the experiments of Louis Pasteur. After that, for the last 200 years, scientists came up with a lot more hypotheses about the emergence of life.
Abiogenesis- Let there be life
Abiogenesis proposes that the first life forms generated on Earth were very simple and became increasingly complex through a gradual process. As a result, trying to trace back evolution to its origin would lead to the last universal common ancestor (LUCA). This is the biological point of view and it can be researched extensively on our planet, where life emerged about 3.5-3.8 billion years ago. The Oparin-Haldane theory supports abiogenesis in the presence of an external energy source, like solar ultraviolet radiation and electric discharges in a reducing atmosphere, but it lacks the precise mechanism that led to the assembly of the first protocell (6). In addition, Oparin assumes that the first life forms were heterotrophs resulting in the heterotroph hypothesis for the evolution of the basic metabolic processes.
A few decades later, the Miller-Urey experiment (7) tested this hypothesis and proved that amino acids, one of the building blocks of biology, could be produced from a mixture of reducing gases (CH4, NH3,H2O and H2). However, it takes multiple steps to synthesise nucleotides and hence RNA, leading to the conclusion that the process specified in the experiment could not be a sufficient mechanism for the origin of life. Additionally, the assumption that the specific gas mixture used in their experiment was a realistic primitive atmosphere is questionable, as it is considered too reducing nowadays (8). As a result, further prebiotic systems chemistry were developed(9), in an attempt to identify any missing intermediates which provide a chemical pathway to biology.
Nevertheless, the theory points out that life evolved through a bottom-up adaptive design method (10), starting from small atoms and molecules that gradually formed larger chains to form informational biopolymers by autocatalysis and eventually cells.
A bottom-up design method is the amalgamation of systems to give rise to more complex systems. Although there has been tremendous progress over the last 30 years, there are certainly gaps in the knowledge of the precise mechanisms. Adam Smith's famous invisible hand could be applied and explain the transition between these non-equilibrium phases. In other words, cellular formation would be part of “the unintended global benefits of individual self-interested actions”. But who governs those actions in the microscale?
It cannot be free will on that level, so it must be the four fundamental forces and scientific laws, specifically the second law of thermodynamics, which states that total entropy increases in an irreversible process.
In the future, the adoption of artificial intelligence software is going to revolutionise process engineering, giving us the freedom of not having to choose a specific synthetic pathway during design. As a next generation chemical engineer, I wouldn't necessarily look for a sequence of mechanisms, as long as the output satisfies the design specifications and I have sufficient information regarding the system’s thermodynamics and kinetics, allowing us to reach the global optimisation point of a process much faster.
In my opinion, the evolution of chemical engineering should inherit more autonomous biological systems design principles, where individual functions are performed independently without the need for centralised permission, just like I don't consciously think about my body's essential metabolic pathways for survival.
Eventually, the origin of life question would be solved more efficiently by using an AI model to simulate it, without any human intervention in the decision-making of the chemical processes. Molecular models explore the dynamics in a system, recording the structural and thermodynamic properties of matter. The main branches of computational algorithms used in molecular simulations are molecular dynamics and Monte Carlo. Although “all models are wrong”, there are techniques like coarse graining which develop physically accurate models of minimal computational complexity (3). Perhaps the main limitation of simulations are the shorter time scales compared to many biological changes.
Having said all that, the research done by the Szostak lab in the last three decades with regards to replicating vesicles, replicating nucleic acids and the role of peptides in prebiotic chemistry is remarkable and a lot easier to understand because of the added animations, which graphically illustrate their published papers. Recently, a kinetic model of nonenzymatic RNA polymerisation was published (11) and even though it does not solve the mystery yet, it brings us one step closer to the answer, implying that it is perfectly feasible to find the precise mechanism with this method. At the same time, it is not practical to focus only on one problem independently in every scenario. The two approaches should be used complementary, where lab experiments would verify any potential breakthroughs achieved by an AI model.
The main reason I strongly believe that an AI model could deal with such a highly complex problem is its expanded capacity and flexibility to treat a dynamic system with carbon, energy and water cycles. Designing a model with an adaptive variable simulation algorithm that considers the evolution of the system would be the goal in the origin field. The observed population of chemical clusters all trying to survive would lead to the conclusion that evolution is not only a trait of living organisms and that LUCA was not one cell, but many with distinguishable variations.
References
- Palmer JC, Debenedetti PG. Recent advances in molecular simulation: A chemical engineering perspective. AIChE Journal. 2015;61(2):370-83.
- Smith E, Morowitz HJ. The Origin and Nature of Life on Earth: The Emergence of the Fourth Geosphere. Cambridge: Cambridge University Press; 2016.
- Sweatman M. Giant SALR cluster reproduction, with implications for their chemical evolution. Sweatman , M 2017 , ' Giant SALR cluster reproduction, with implications for their chemical evolution ' Molecular Physics DOI: 101080/0026897620171406164. 2017.
- Tirard S. J. B. S. Haldane and the origin of life. Published by the Indian Academy of Sciences. 2017;96(5):735-9.
- Miller SL. A Production of Amino Acids under Possible Primitive Earth Conditions. Science. 1953;117(3046):528-9.
- Ferus M, Pietrucci F, Saitta AM, Knížek A, Kubelík P, Ivanek O, et al. Formation of nucleobases in a Miller-Urey reducing atmosphere. Proceedings of the National Academy of Sciences of the United States of America. 2017;114(17):4306.
- Islam S, Powner MW. Prebiotic Systems Chemistry: Complexity Overcoming Clutter. Chem. 2017;2(4):470-501.
- Pereto J. Out of fuzzy chemistry: from prebiotic chemistry to metabolic networks. Chemical Society Reviews. 2012;41(16):5394-403.
- Walton T, Szostak JW. A Kinetic Model of Nonenzymatic RNA Polymerization by Cytidine-5′-phosphoro-2-aminoimidazolide. Biochemistry. 2017;56(43):5739-47.
Note: Steem has re-numbered the reference list starting from 1, but the correct list starts at reference 3 and finishes at 11. Thanks again for reading.