Proteins are fundamental macromolecules accomplish various cellular functions of living systems, including hormonal signaling, structural, transport and reaction catalyst regulation. These polymers correspond to linear chains composed of an amino acid sequence which determines their three-dimensional structure. The protein topology corresponds to the spatial arrangement of the polypeptide chain and its elements of secondary structure, such as alpha helix and beta sheets. Alpha helix and beta sheets are arranged in domains like alpha/beta topology that consists of a repetition of the alpha/beta/alpha motif, leaving the outer surface formed by alpha propellers that are packed against a central skeleton of beta sheets. Topology is key to understanding protein folding, due to the clues it gives about possible conformations and those that are not.
So, how do proteins move from a linear state to a complex three-dimensional structure? According to Branden and Tooze, for a protein to be functional it must be correctly folded, that is, it must find its "native conformation." If the amino acid sequence determines its structure (Anfinsen's dogma), then it should also direct it to native conformation.
One of the most exciting aspects of proteins is the ability of their polypeptide chain to self-roll or fold to generate three-dimensional structures, and even more curious, to knot; this is why it becomes even more complicated when they present topologies that were thought impossible. These types of proteins form true knots, which can be simple or complex such as nautical knots (Virnau et al., 2011). The knots were proposed more than 30 years ago (Crippen G, 1974; Liu L. 1976 and Wang H, 1996). The first studies were made more than 15 years ago in carbonic anhydrase B, where a superficial knot was found, which was at one end of the molecule and is easily deployed (Mansfield M, 1994). Over the years, knots have been seen to be scarce (1% of the PDB), although there are families such as SpoU 2' O-ribose methyl transferase and carbon dioxide anhydrases (Murzin A, 1995; Taylor W, 2000).
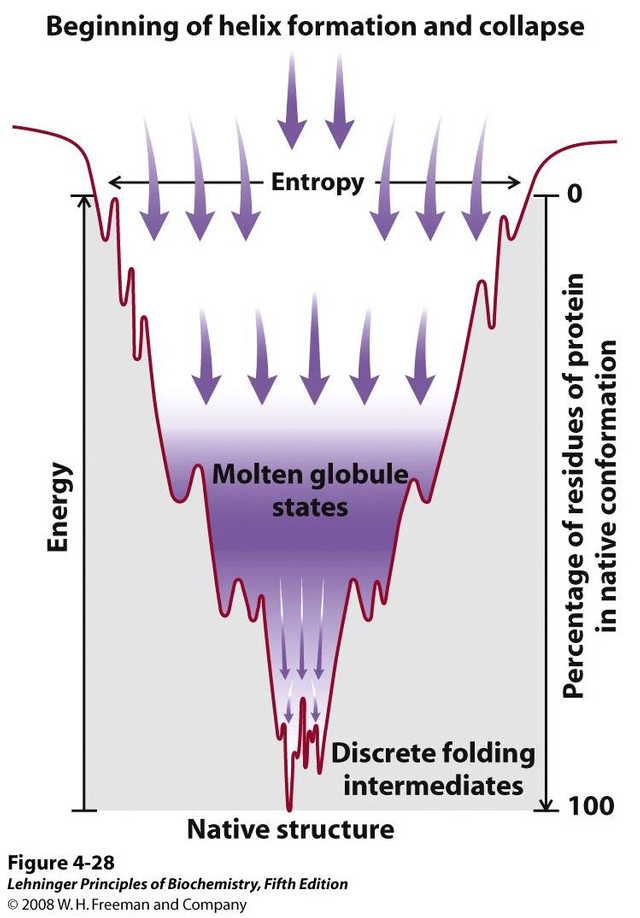
There are many folding theories that explain how they move from a fully deployed state to a complex tertiary structure through collapses, rearrangements and intermediary formation (Dagget V. Y Fersht A., 2003 and Rousseau F., 2002). Multiple models have been proposed over the years, but recently a unified model of protein folding based on the effective energy surface has emerged. These new models have evolved and been simplified and are illustrated by the folding funnel model. This model integrates the energetic landscape, where the thermodynamic behavior of the transformation of the set of molecules deployed to a predominantly native state is described. This integration of models proposes that there is no single folding route, but rather that there are different routes that arrive at a single conformation. These models follow the principle of "minimal frustration" (Onuchic 1997; and Bryngelson, 1989) where it is considered that the structures have evolved in such a way that sequences are selected allowing to favor the formation of the native state in a low energy structure, which allows minimizing interactions that come into conflict with the native structure (Monroe and cols, 2010).
One interesting model to study knotted proteins is the protein YibK, member of the family? /? methyltransferase whose topology is "trefoil" type or 3 1 (Figure 1). In studies where it has been denatured chemically and through techniques such as fluorescence and mutagenesis of residues involved in the core of the node of this methyltransferase do not influence the knotted topology of the protein. Also, Mallan A. et al (2010) by means of chemical denaturation, were able to determine that binding the terminal ends of YibK with the ThiS protein (Mallan A. et al, 2008) had a very small influence on the folding kinetics of this protein, asserting also that the formation of the node involves cooperation with folding and channels the protein to the native state. For this reason, it was concluded that the formation of the knot and the collapse of the protein will be carried out independently.

References (selected)
Benítez-Paez, Villaroya,Douthwaite, Galbadon (2010). YibK is the 29-O methyltransferase TrmL that modifies the wobble nucleotide in Escherichia coli tRNALeu isoacceptors. RNA , 16:2131–2143.
Borgia, Williams and Clarke (2008). Single-Molecule Studies of Protein Folding. Annu. Rev. Biochem, 77:101–25.
Bornschlogl, Anstrom, Mey, Dzubiella, Rief and Forest (2009). Tightening the Knot in Phytochrome by Single-Molecule Atomic Force Microscopy. Biophysical Journal, Volume 96, 1508–1514.
Cabrita, Dai and Bottomley (2006). A family of E. coli expression vectors for laboratory scale and high throughput soluble protein production. BMC Biotechnology, 1472-6750.
Cecconi, Shank, Bustamante, Marqusee (2005). Direct Observation of the Three-State Folding of a Single Protein Molecule. Science, Vol 309, 2057-2060.
Cecconi, Shank, Marqusee and Bustamante (2011). DNA Molecular Handles for Single-Molecule Protein-Folding Studies by Optical Tweezers.Methods Mol Biol. 749:255-71.
Fersht. Structure and Mechanism in Protein Science. W.H. Freeman and Company, New
York, 1999.Jackson SE (1998), How do small single-domain proteins fold?. Fold Des.3(4):R81-91.
Mallam and Jackson (2005). Folding Studies on a Knotted Protein. J. Mol. Biol. 346, 1409–1421.
Mallam, Morris, Jackson (2008).Exploring knotting mechanisms in protein folding. Mallam, Morris, Jackson. PNAS, vol. 105, no. 48, 18740–18745.
Mallam, Onuoha, Grossmann, Jackson (2008). Knotted Fusion Proteins Reveal Unexpected
Possibilities in Protein Folding. Molecular Cell 30, 642–648.Mallam, Rogers, and Jackson (2010). Experimental detection of knotted conformations. PNAS, vol. 107, no. 18, 8189–8194.
Onuchic. Luthey-Schulten, and Wolynes (1997). Theory of protein folding: The energy
landscape perspective. Annu. Rev. Phys. Chem, 48:545–600.
Stols, Gu, Dieckman, Raffen, Collart and Donnelly (2002). A New Vector for High-Throughput, Ligation-Independent Cloning Encoding a Tobacco Etch Virus Protease Cleavage Site. Protein Expression and Purification 25, 8–15.
Virnau, Mallam and Jackson(2011). Structures and folding pathways of topologically knotted proteins.J. Phys.: Condens. Matter 23.
Congratulations @miquemau! You have received a personal award!
Click on the badge to view your Board of Honor.
Do not miss the last post from @steemitboard:
SteemitBoard World Cup Contest - Brazil vs Belgium
Participate in the SteemitBoard World Cup Contest!
Collect World Cup badges and win free SBD
Support the Gold Sponsors of the contest: @good-karma and @lukestokes
Downvoting a post can decrease pending rewards and make it less visible. Common reasons:
Submit
Congratulations @miquemau! You received a personal award!
You can view your badges on your Steem Board and compare to others on the Steem Ranking
Vote for @Steemitboard as a witness to get one more award and increased upvotes!
Downvoting a post can decrease pending rewards and make it less visible. Common reasons:
Submit