
Cells differ in the proteins they express.
The same cell may express different proteins at different times in its life, and in multicellular organisms, different types of cells typically express different suites of proteins. Given that all cells in an organism have the same set of genes, how do these differences in protein expression arise? Part I outlines the history of the original experiments done by Jacques Monod and François Jacob in the late 1950s and early 1960s using bacteria to discover the basic mechanisms of gene regulation.
Though we have discussed cell specialization, we have not examined what makes cells different from each other.
Differences in cell function often produce different cell shapes, but the critical difference is the different protein mix each type of cell contains. Though many thousands of proteins are coded for in a typical eukaryotic cell, most cells contain only a fraction of that number at any one time. This is the case even in single-celled bacteria. This situation makes sense because making proteins is expensive and many proteins do not store well. This leads to the question of how cells control protein production. We know that making proteins requires the transcription of DNA into mRNA, then the translation of mRNA into proteins; thus, we could alternatively ask why all genes in a cell are not always transcribed and translated. The general answer is that cells regulate gene expression by controlling the conditions necessary for transcription and translation.
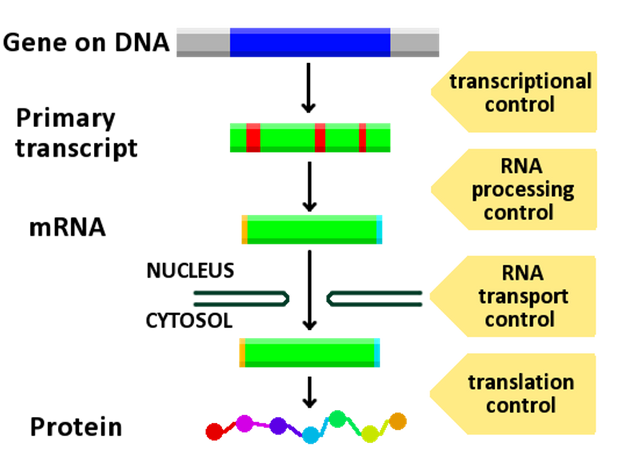
The complexity of gene expression means that there are many possible points where cells can control it.
In addition to transcription and translation, the initial mRNA transcript must be processed before it is translated, and the final polypeptide must often be further modified by enzymes before it is functional. Cells could theoretically control gene expression at any of these points, but the majority of gene regulation happens at the level of transcription—specifically, by controlling mRNA synthesis. This is the case primarily for efficiency; it “costs” cells less to prevent the process from starting in the first place.
Control of transcription mechanisms involves genetic “lock-and-key” mechanisms, which work differently in prokaryotes and eukaryotes.
To a first approximation, the “switch” that turns transcription on and off is a lock-and-key mechanism. The lock is a specific sequence of nucleotide bases on the DNA that is distinct from but often located physically adjacent to the gene it controls. Each gene has one or more of these regulatory regions, which normally occur “upstream” from the gene. The key is usually a protein with the right shape (including not only physical shape but the correct physical and chemical properties of amino acids) to fit the DNA lock. In general, only one protein will bind to a particular regulatory region.
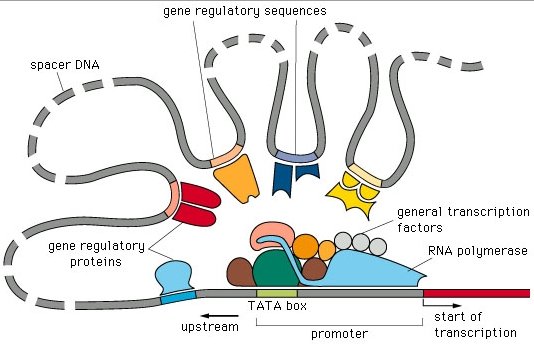
In order to start transcription, RNA polymerase must bind to promoter sites on the DNA molecule. The binding of RNA polymerase to a promoter is not specific; thus, regulatory regions and proteins generally act in two ways to control gene expression.
In prokaryotic cells, regulatory regions typically lie in between promoter sites and genes. Regulatory proteins bound to regulatory regions physically block RNA polymerase from reaching the gene. This represents negative control and is typical of prokaryotic cells.
The other type of control depends on the fact that RNA polymerase cannot always bind efficiently to the promoter by itself. In this case regulatory proteins interacting with regulatory regions affect the ability of RNA polymerase to bind to a promoter. In the most general case, regulatory proteins (called “transcription factors”) facilitate the binding of RNA polymerase, resulting in positive control, though these proteins can have either a positive or negative effect on binding in specific cases.
These types of controls create two contrasts: physical blockage of RNA polymerase versus an effect on its ability to bind to a promoter region on the DNA, and negative control versus positive control.
Though the lock-and-key mechanism for gene regulation can be compared to turning on a car, there are some important differences.
- The “driver” (RNA polymerase) is not selective as to which “car” it chooses. RNA polymerase simply tries to transcribe every gene.
- Some genes will be expressed only if the key is not in the regulatory lock.
- Some genes require a combination of many different keys to be expressed, and some keys must be in their locks, while others must be out.

In the late 1950s and early 1960s, Jacques Monod and François Jacob investigated how gene regulation works in prokaryotes.
The bacteria Escherichia coli (E. coli for short) requires several enzymes to metabolize lactose. Biochemists discovered in the early 20th century that E. coli produces these enzymes only when lactose is available, suggesting that lactose induces enzyme production. This observation led Jacques Monod and François Jacob to the question of how lactose could induce gene expression; they found mutant forms of E. coli that differed in how they expressed lactose-digesting enzymes.
Two kinds of mutants were unable to digest lactose; in one case, an enzyme involved in lactose breakdown itself was defective, while in another case, an enzyme that brings lactose into the cell was defective. These mutations fit into the expected pattern: A mutation in a gene causes a coding error that produces a dysfunctional protein, which cannot do its job.
A third kind of mutant always produced lactose-digesting enzymes, regardless of whether lactose was present or not. This type of mutation produced a gain of constant function rather than a loss of function. Monod and Jacob inferred that this third type of mutant must have a mutation in some protein involved in controlling gene expression, not in the genes coding for the enzymes themselves.
Monod and Jacob’s idea was revolutionary at the time, because it pointed to a protein whose sole function was to regulate the expression of other genes.
In E. coli, the regulatory protein seemed to inhibit enzyme production. After some debate, physicist Leo Szilard prevailed with the idea that the regulatory protein acted as a repressor that normally prevented the production of the lactose-digesting enzymes. Szilard’s model suggested that lactose acts as an inducer by disabling the repressor protein; that is, expression of the lactose-digesting enzymes is under negative control.
Questions to Comment on:
Is turning on a car an example of positive control or negative control?
Why was it surprising to Jacob and Monod to find a mutation of a gene that caused a functional protein that is normally absent in a cell to be produced?
Image Credits -
https://dr282zn36sxxg.cloudfront.net/datastreams/f-d%3A0b7982437d893524a649c2245f4cd034d1c2776094e1c39a72b5f627%2BIMAGE%2BIMAGE.1
https://upload.wikimedia.org/wikipedia/commons/1/11/Gene_expression_control.png
http://pbil.univ-lyon1.fr/members/sagot/htdocs/team/projects/chromo_net/images/reg.jpg
http://i.f1g.fr/media/ext/805x525/www.lefigaro.fr/medias/2013/04/21/PHOb40f5994-aac4-11e2-9c91-a3a3a94a97f7-805x525.jpg
Bird [Giphy](https://giphy.com/gifs/blue-5lDik5jPpRw2Y)

BTC: 1JQWNxziArfaCtxsP3tUQ4VSTBY9jresmt 



The paper I describe in my latest IGMS column uses this exact principle, which I just referred to as epigenetic without going into the mechanism. I'm going to share this post with my readers to help them out with the details.
http://www.intergalacticmedicineshow.com/cgi-bin/mag.cgi?do=columns&vol=randall_hayes&article=010
Downvoting a post can decrease pending rewards and make it less visible. Common reasons:
Submit