An alternative to the seven-to-nine tubulin patches in Craddock, Tuszynski, and Hameroff's 2012 paper (link), here with six tubulin patches instead, with possible bit states. I have remixed their work, images, and similar, to show a better model.
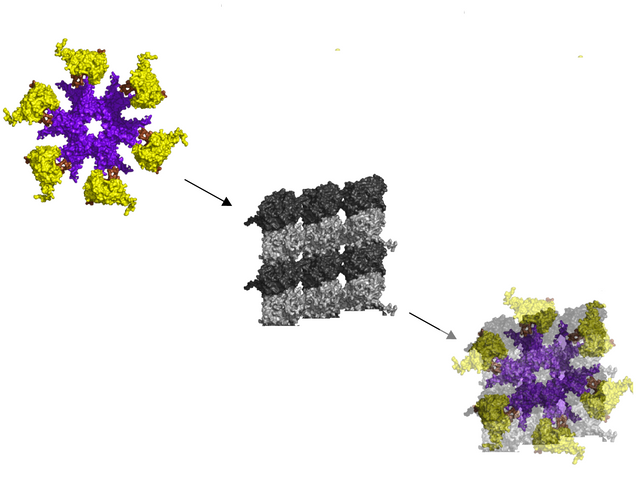
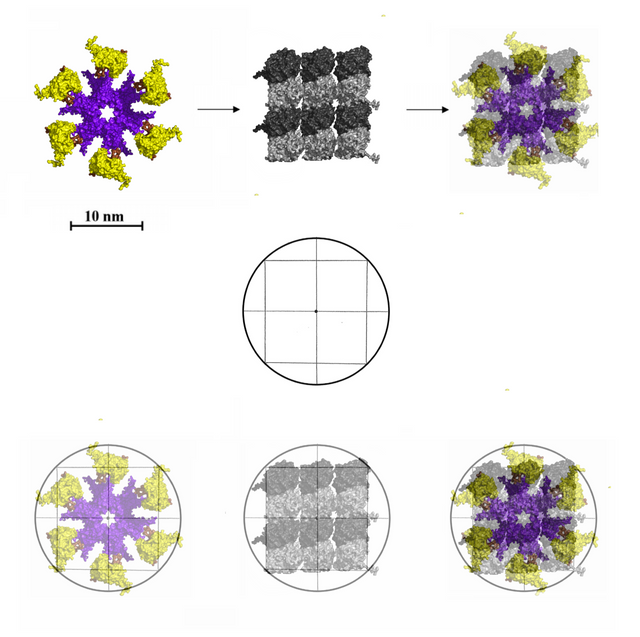
The new model, a six tubulin fit for CAMKII on MT lattices
CAMKII from below, six arms with kinase domains, fitted onto a six tubulin lattice, with 64 possible bit-states per CaMKII-MT byte.
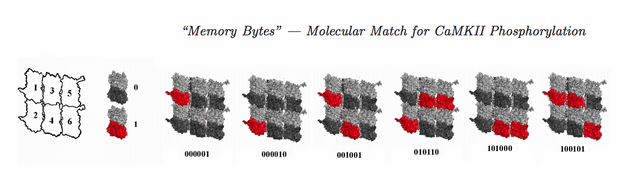
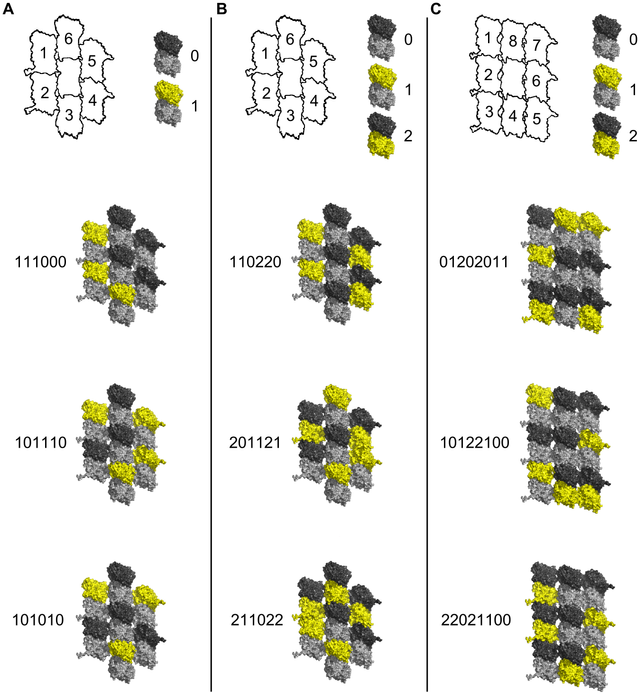
Memory states of a microtubule lattice, six tubulin (from CAMKII six-arm fit) with phosphorylation states. Six out of 26 (64) possible states are shown.
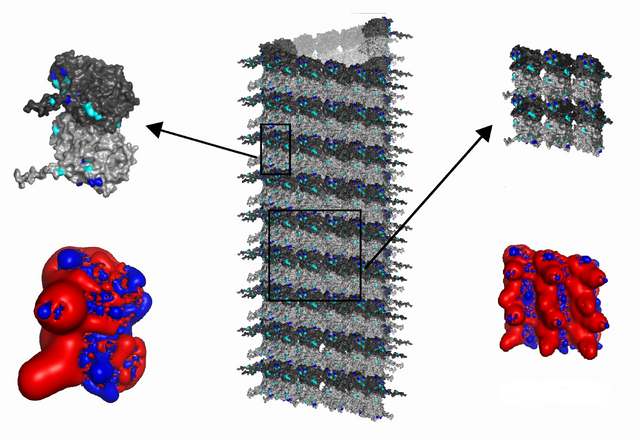
The tubulin dimers are 8 nm long and 4.5 nm wide, giving a 3 x 2 lattice field a height of 16 nm and a width of 13.5 nm, and each column of three will have two spaces between tubulin while only one space for the row of two, so the 3 x 2 tubulin field approximates a square, and the microtubule is also curved which means CAMKII has to bend itself, shortening the distance between kinase domains along the width of the MT lattice. In other words, if the MT is adapted to a 2D projection then the lattice field is stretched along its width, forming a perfect square.
The 20 nm CAMKII bends along the width of the 3 x 2 tubulin field, to s = rθ = 16.37 nm, the diameter s of CAMKII along the width of the tubulin field is 5/6th of the 20 nm diameter along the length of the tubulin field.
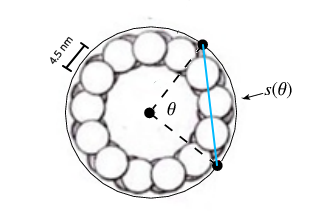
For the CAMKII write-heads, the tubulin field is stretched with 1.2x along the curved width of the microtubule and unchanged along the length of the microtubule, giving a width of roughly 16 nm, the same as the length of 16 nm, from two 8 nano meter tubulin dimers.
A comparison with Craddock, Tuszynski, and Hameroff's model
In Cytoskeletal Signaling: Is Memory Encoded in Microtubule Lattices by CaMKII Phosphorylation? from 2012, the authors suggest a seven tubulin fit as well as a nine tubulin fit.
In their model with 7-tubulin lattice fields, the middle tubulin is an "address" dimer unavailable for phosphorylation.
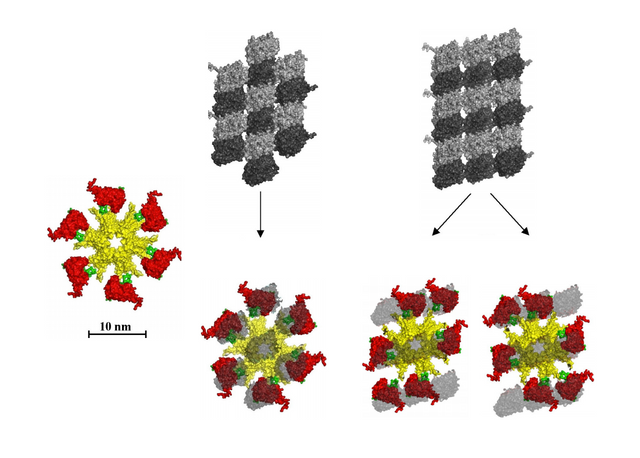
To me, it seems like a six tubulin fit makes more sense, if CAMKII is an intermediary that writes 6 "bits" onto microtubule lattices, a "byte" in the MT lattice as a memory storage device, and CAMKII has six arms with kinases, and seems to fit onto a six tubulin site, seems like a better model.
Below is Craddock, Tuszynski, and Hameroff's model for memory states on a seven and nine tubule field, to be compared to my model with a six tubule field.
With a 7-tubulin MT "byte", is not the three rows 8 nm x 3 = 24 nm, outside the reach of a 20 nm CAMKII?
Tubulin monomers are 4 nm long and dimers 8 nm, so the 8 nm x 3 = 24 nm 7-tubulin lattice "byte" is one tubulin monomer, one bit-state, outside the reach of CAMKII.
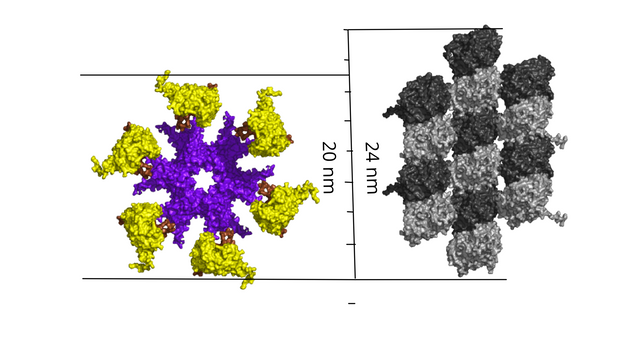
The 3 x 3 (9) tubulin lattice field is also one tubulin monomer (one bit-state) too high, 3 x 8 nm = 24 nm, with a width of 3 x 5.8 nm (space a tubulin filament occupies, tubulin are 4.5 nm) = 17.4 nm, if adjusted for the curvature (if CAMKII "sees" a flat surface) it is 15.9 nm wide, almost the same width as a two row lattice "patch" is high, 8 nm x 2 = 16 nm.
3 x 2 tubulin lattice fields as a MT byte
The idea here is that the MT "byte" is not a 15.9 nm x 24 nm rectangle, but instead a perfect square, 4/5th the width of CAMKIIs 20 nm diameter, and a 3 x 2 tubulin "patch" fits that perfectly if curvature is adjusted for, CAMKII "sees" a 15.9 x 16 nm lattice patch.
First its great to see this topic again and its good that looks you got serious information about camkii..and is it possible what are yoi saying camkii.
Downvoting a post can decrease pending rewards and make it less visible. Common reasons:
Submit
Downvoting a post can decrease pending rewards and make it less visible. Common reasons:
Submit